Getting insights from data is exciting. So let's see how well I can cover this topic in a single article.
In this article, I assume you have data in a single CSV file. If you have
multiple CSV files, you can merge
them similar to SQL JOIN statements.
Prerequesites
- Python 3
- Pandas (
pip install pandas) - Matplotlib (
pip install matplotlib) - Seaborn (
pip install seaborn)
Loading the data
I load the data as a Pandas Dataframe first. I name this dataframe df.
If the CSV file is too big for RAM, I use the command line tool head to crop
the data. Dask is another option, but I
never actually used it.
I use the titanic training data as an example:
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
import pandas as pd
df = pd.read_csv("train.csv")
How many datapoints do we have?
print("Total number of datapoints: {datapoints}".format(datapoints=len(df)))
shows we have 891 data points in total. Not too much.
Which columns do we have?
print(df.describe())
reveals
PassengerId Survived Pclass Age SibSp \
count 891.000000 891.000000 891.000000 714.000000 891.000000
mean 446.000000 0.383838 2.308642 29.699118 0.523008
std 257.353842 0.486592 0.836071 14.526497 1.102743
min 1.000000 0.000000 1.000000 0.420000 0.000000
25% 223.500000 0.000000 2.000000 NaN 0.000000
50% 446.000000 0.000000 3.000000 NaN 0.000000
75% 668.500000 1.000000 3.000000 NaN 1.000000
max 891.000000 1.000000 3.000000 80.000000 8.000000
Parch Fare
count 891.000000 891.000000
mean 0.381594 32.204208
std 0.806057 49.693429
min 0.000000 0.000000
25% 0.000000 7.910400
50% 0.000000 14.454200
75% 0.000000 31.000000
max 6.000000 512.329200
Ok, so we have:
PassengerId: 891 non-nan values. Just an identifier.Survived: 891 non-nan values. I guess only true / false?Pclass: 891 non-nan values. Min=1 and max=3, I guess of type int and thus three classes of passengers?Age: only 714 non-nan values. 0.42 is a weird value ... something like 5 months. And a maximum of 80. So likely in years.- SibSp: According to Kaggle docs: '# of siblings / spouses aboard the Titanic'
- Parch: According to Kaggle docs: '# of parents / children aboard the Titanic'
- Fare: Not sure which unit would make sense here.
Pandas df.describe() is not powerfull enough. So I wrote my own:
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
"""Exploratory data analysis."""
# core modules
import logging
import sys
# 3rd party module
import pandas as pd
logging.basicConfig(
format="%(asctime)s %(levelname)s %(message)s",
level=logging.DEBUG,
stream=sys.stdout,
)
def main(csv_filepath):
"""Exploratory data analysis for the Titanic dataset."""
# Read data
dtype = {
"PassengerId": "str",
"Embarked": "category",
"Survived": "category",
"Pclass": "category",
"Sex": "category",
"SibSp": "uint8",
"Parch": "uint8",
}
df = pd.read_csv(csv_filepath, dtype=dtype)
describe_pandas_df(df, dtype=dtype)
def describe_pandas_df(df, dtype=None):
"""
Show basic information about a pandas dataframe.
Parameters
----------
df : Pandas Dataframe object
dtype : dict
Maps column names to types
"""
if dtype is None:
dtype = {}
print("Number of datapoints: {datapoints}".format(datapoints=len(df)))
column_info = {"int": [], "float": [], "category": [], "other": []}
float_types = ["float64"]
integer_types = ["int64", "uint8"]
other_types = ["object", "category"]
column_info_meta = {}
for column_name in df:
column_info_meta[column_name] = {}
counter_obj = df[column_name].groupby(df[column_name]).count()
value_list = list(counter_obj.keys())
value_count = len(value_list)
is_suspicious_cat = (
value_count <= 50
and str(df[column_name].dtype) != "category"
and column_name not in dtype
)
if is_suspicious_cat:
logging.warning(
"Column '{}' has only {} different values ({}). "
"You might want to make it a 'category'".format(
column_name, value_count, value_list
)
)
top_count_val = counter_obj[value_list[0]]
column_info_meta[column_name]["top_count_val"] = top_count_val
column_info_meta[column_name]["value_list"] = value_list
column_info_meta[column_name]["value_count"] = value_count
if df[column_name].dtype in integer_types:
column_info["int"].append(column_name)
elif df[column_name].dtype in float_types:
column_info["float"].append(column_name)
elif str(df[column_name].dtype) == "category":
column_info["category"].append(column_name)
elif df[column_name].dtype in other_types:
column_info["other"].append(column_name)
else:
print(
"!!! describe_pandas_df does not know type '{}'".format(
df[column_name].dtype
)
)
column_name_len = max(len(column_name) for column_name in df)
print("\n## Integer Columns")
print(
"{column_name:<{column_name_len}}: Non-nan mean std min 25% "
" 50% 75% max".format(
column_name_len=column_name_len, column_name="Column name"
)
)
for column_name in column_info["int"]:
print(
"{column_name:<{column_name_len}}: {non_nan:>7} "
"{mean:0.2f} {std:>4.2f} "
"{min:>4.0f} {q25:>4.0f} {q50:>4.0f} {q75:>4.0f} {max:>4.0f}".format(
column_name_len=column_name_len,
column_name=column_name,
non_nan=sum(df[column_name].notnull()),
mean=df[column_name].mean(),
std=df[column_name].std(),
min=df[column_name].min(),
q25=df[column_name].quantile(0.25),
q50=df[column_name].quantile(0.50),
q75=df[column_name].quantile(0.75),
max=max(df[column_name]),
)
)
print("\n## Float Columns")
print(
"{column_name:<{column_name_len}}: Non-nan mean std min "
"25% 50% 75% max".format(
column_name_len=column_name_len, column_name="Column name"
)
)
for column_name in column_info["float"]:
print(
"{column_name:<{column_name_len}}: {non_nan:>7} "
"{mean:5.2f} {std:>4.2f} "
"{min:>5.2f} {q25:>5.2f} {q50:>5.2f} {q75:>5.2f} {max:>5.2f}".format(
column_name_len=column_name_len,
column_name=column_name,
non_nan=sum(df[column_name].notnull()),
mean=df[column_name].mean(),
std=df[column_name].std(),
min=df[column_name].min(),
q25=df[column_name].quantile(0.25),
q50=df[column_name].quantile(0.50),
q75=df[column_name].quantile(0.75),
max=max(df[column_name]),
)
)
print("\n## Category Columns")
print(
"{column_name:<{column_name_len}}: Non-nan unique top (count) "
"rest".format(column_name_len=column_name_len, column_name="Column name")
)
for column_name in column_info["category"]:
# print(df[column_name].describe())
rest_str = str(column_info_meta[column_name]["value_list"][1:])[:40]
print(
"{column_name:<{column_name_len}}: {non_nan:>7} {unique:>6} "
"{top} ({count}) {rest}".format(
column_name_len=column_name_len,
column_name=column_name,
non_nan=sum(df[column_name].notnull()),
unique=len(df[column_name].unique()),
top=column_info_meta[column_name]["value_list"][0],
count=column_info_meta[column_name]["top_count_val"],
rest=rest_str,
)
)
print("\n## Other Columns")
print(
"{column_name:<{column_name_len}}: Non-nan unique top (count)".format(
column_name_len=column_name_len, column_name="Column name"
)
)
for column_name in column_info["other"]:
rest_str = str(column_info_meta[column_name]["value_list"][1:])[:40]
print(
"{column_name:<{column_name_len}}: {non_nan:>7} {unique:>6} "
"{top} ({count})".format(
column_name_len=column_name_len,
column_name=column_name,
non_nan=sum(df[column_name].notnull()),
unique=len(df[column_name].unique()),
top=column_info_meta[column_name]["value_list"][0],
count=column_info_meta[column_name]["top_count_val"],
)
)
def get_parser():
"""Get parser object for exploratory data analysis."""
from argparse import ArgumentParser, ArgumentDefaultsHelpFormatter
parser = ArgumentParser(
description=__doc__, formatter_class=ArgumentDefaultsHelpFormatter
)
parser.add_argument(
"-f",
"--file",
dest="filename",
help="read this csv file",
metavar="FILE",
required=True,
)
return parser
if __name__ == "__main__":
args = get_parser().parse_args()
main(args.filename)
which gives:
Number of datapoints: 891
## Integer Columns
Column name: Non-nan mean std min 25% 50% 75% max
SibSp : 891 0.52 1.10 0 0 0 1 8
Parch : 891 0.38 0.81 0 0 0 0 6
## Float Columns
Column name: Non-nan mean std min 25% 50% 75% max
Age : 714 29.70 14.53 0.42 20.12 28.00 38.00 80.00
Fare : 891 32.20 49.69 0.00 7.91 14.45 31.00 512.33
## Category Columns
Column name: Non-nan unique top (count) rest
Survived : 891 2 0 (549) ['1']
Pclass : 891 3 1 (216) ['2', '3']
Sex : 891 2 female (314) ['male']
Embarked : 889 4 C (168) ['Q', 'S']
## Other Columns
Column name: Non-nan unique top (count)
PassengerId: 891 891 1 (1)
Name : 891 891 Abbing, Mr. Anthony (1)
Ticket : 891 681 110152 (3)
Cabin : 204 148 A10 (1)
This is much more useful. In the process, I created a dictionary dtypes which
tells pandas as which type it should load the columns.
Also interesting:
df.groupby(df["foo"]).count()
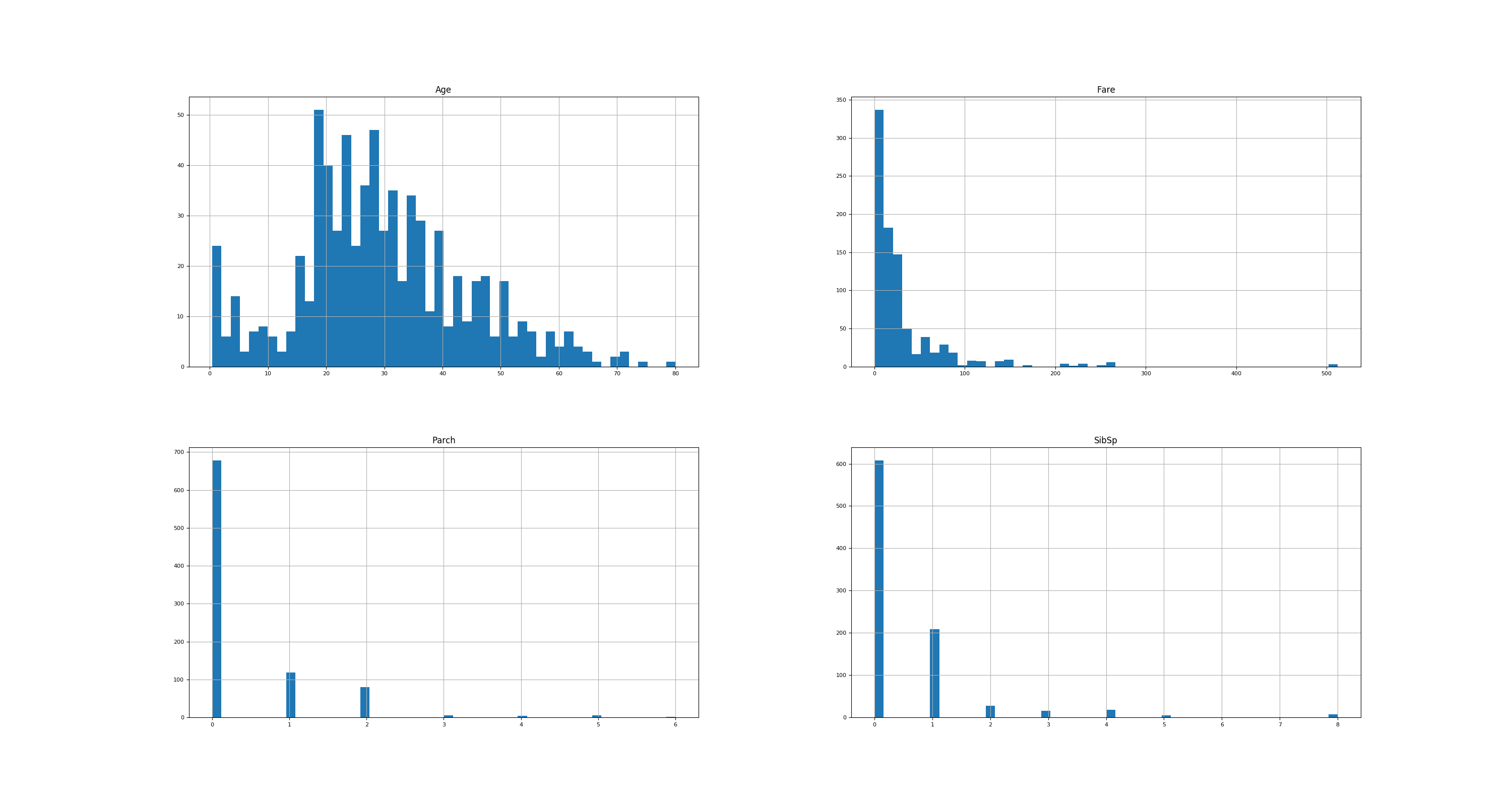
Show single-feature distribution
# Show histograms
numeric_types = ["float64", "int64", "uint8"]
numerical_features = df.select_dtypes(include=numeric_types)
numerical_features.hist(figsize=(30, 16), bins=50, xlabelsize=8, ylabelsize=8)
plt.savefig("titanic-histograms.png")
plt.show()
gives
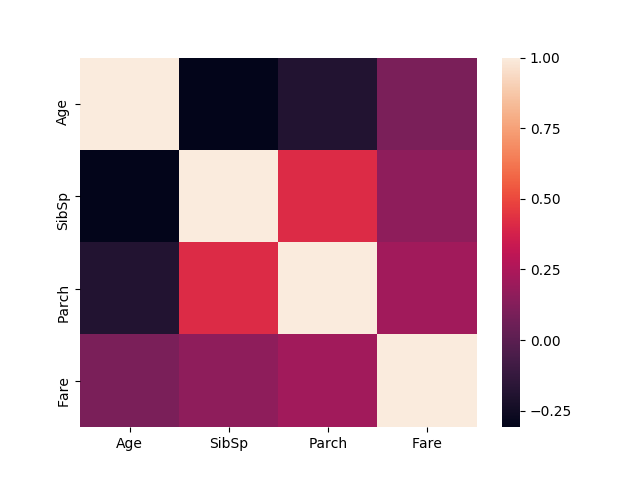
Find correlations
# Show correlations
import seaborn as sns
corr = numerical_features.corr()
sns.heatmap(corr)
plt.savefig("titanic-correlation.png")
plt.show()
gives
Joint plots
See also
- Chloe Mawer, Jonathan Whitmore - Exploratory data analysis in python - PyCon 2017